How DeepMind Solves 3D Structures of Proteins?
Scientists were trying to solve this one problem for 50 years. And we have the privilege to live in an era in which it was solved. Scientists can now computationally determine the 3D structures of proteins.

I almost can’t believe that last week, almost simultaneously as I hit send on my last issue, some ground-shaking news arrived. One word: DeepMind.
So, what happened?
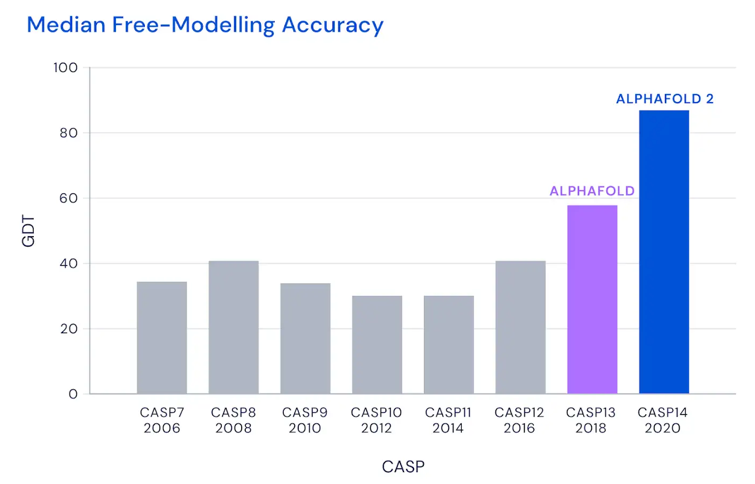
There’s this thing called CASP, short for “Critical Assessment of Structure Prediction”, which is a challenge for teams to predict protein structure. DeepMind and their AI system called AlphaFold, that’s developed by Google, managed to significantly outperform 100 other teams. This isn’t the first time this happened. They also won in 2018, but the big deal this year is that the AlphaFold scored 92.4 on the accuracy score.
Read the rest of the news article from Nature
But this newsletter isn’t just about breaking news and summarising. So in the next few paragraphs, I’ll attempt to paint you (and myself) a picture of what they achieved. Along with the resources I found on the way.
How does it work?
Knowing the 3D structure of proteins is very useful (you don’t say). It can tell us everything about them - what they do in cells, where they bind to, how you can target them, etc. Today, protein structures are mostly determined using X-ray crystallography and NMR - kind of reverse-engineering. We isolate a protein and you determine its structure. This is expensive and time-consuming, which results that only a fraction of protein structures are known in detail.
”These techniques, as well as newer methods like cryo-electron microscopy, depend on extensive trial and error, which can take years of painstaking and laborious work per structure, and require the use of multi-million dollar specialised equipment.” - DeepMind
AlphaFold, and AI systems in general, turn this on its head. We don’t need highly specialised equipment and years of experimental work. Furthermore, we almost determine the protein structure “proactively”, not “reactively”. In an ideal world, we would find a new amino acid sequence and determine what type of protein it encodes and how this protein “looks like”.
AlphaFold is a deep-learning model, which means it was developed to learn to recognise how proteins fold by itself. Essentially, the input is the amino acid sequence and a known protein structure. After lots of data, it should figure out a way to predict how an amino acid sequence might fold into a 3D protein structure. Genomics and proteomics are perfect for this as lots of data is available. Given that there are about 10^300 possible structures for every amino acid sequence, this greatly speeds up the process.
This is not even the only initiative to solve the so-called “protein folding problem”. IBM’s BlueGene and FoldIt initiative are just two of them. Another awesome project is FoldingAtHome, which connects PCs from around the world to help find new therapies for COVID-19.
Determining protein structure is a 50-year-old problem, which could now even be considered solved. CASP determined that an accuracy of more than 90 can be considered the solution. The measure of accuracy is Global Distance Test (GDT), which “can be approximately thought of as the percentage of amino acid residues (beads in the protein chain) within a threshold distance from the correct position”. For AlphaFold, who achieved an accuracy of 92.4GDT, this means their solution is within 1.6 Angstroms (one atom) of the correct experimental solution.
The whole point of CASP is to give research teams protein structures to solve, whose structures were only recently experimentally validated. The teams solve the problem blindly and receive the results later. This was also the case when the teams had to determine the structure of Orf8, a COVID-19 protein. AlphaFold of did this with significant accuracy.
Below is an example of the experimental result and the computational prediction of AlphaFold for two proteins.
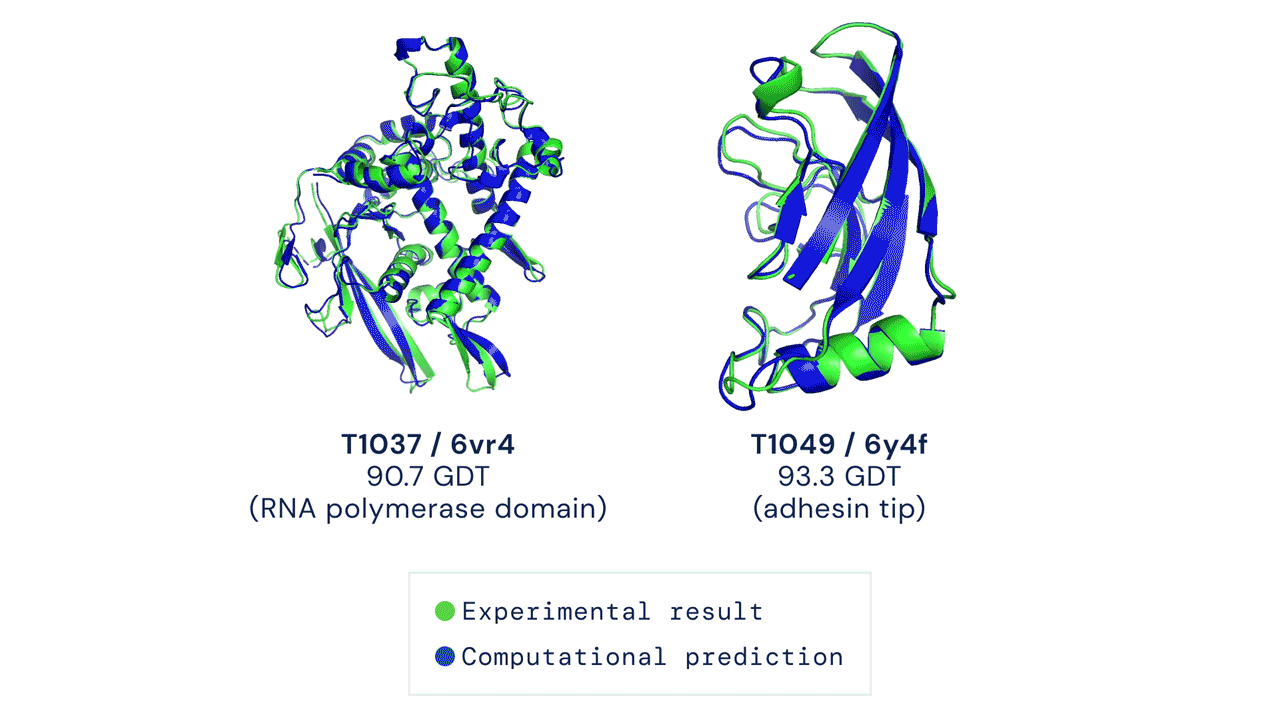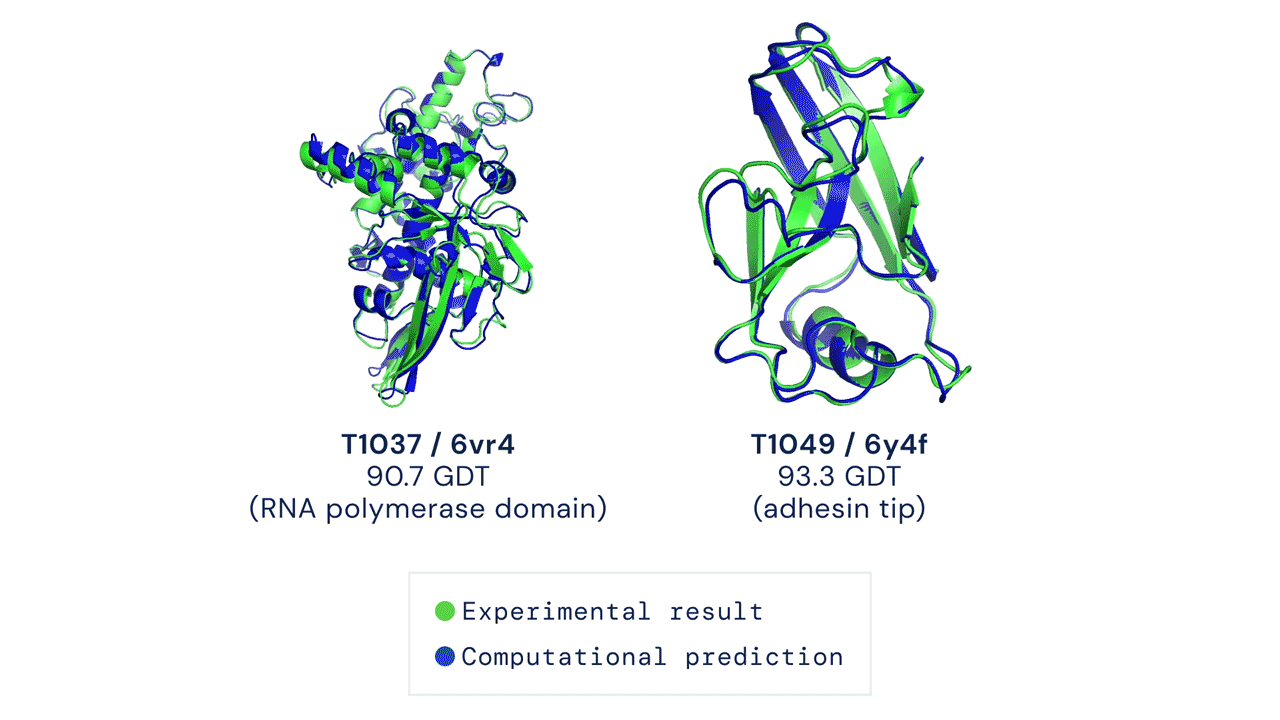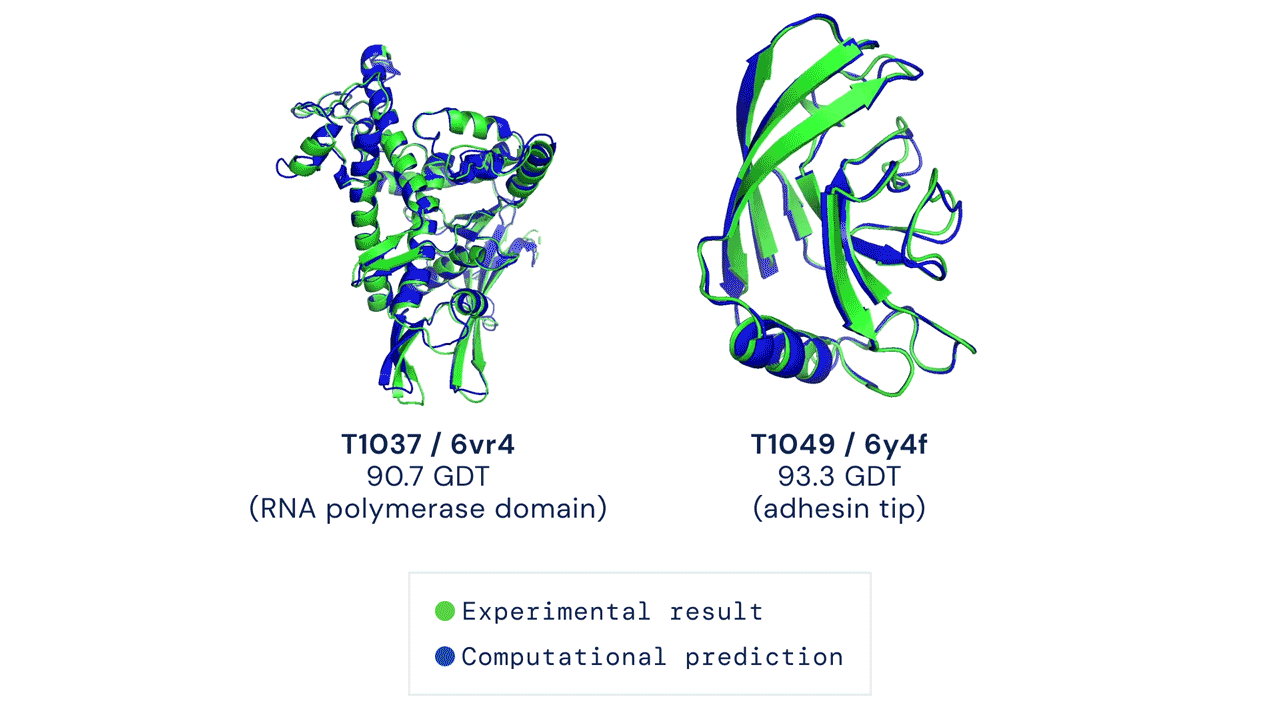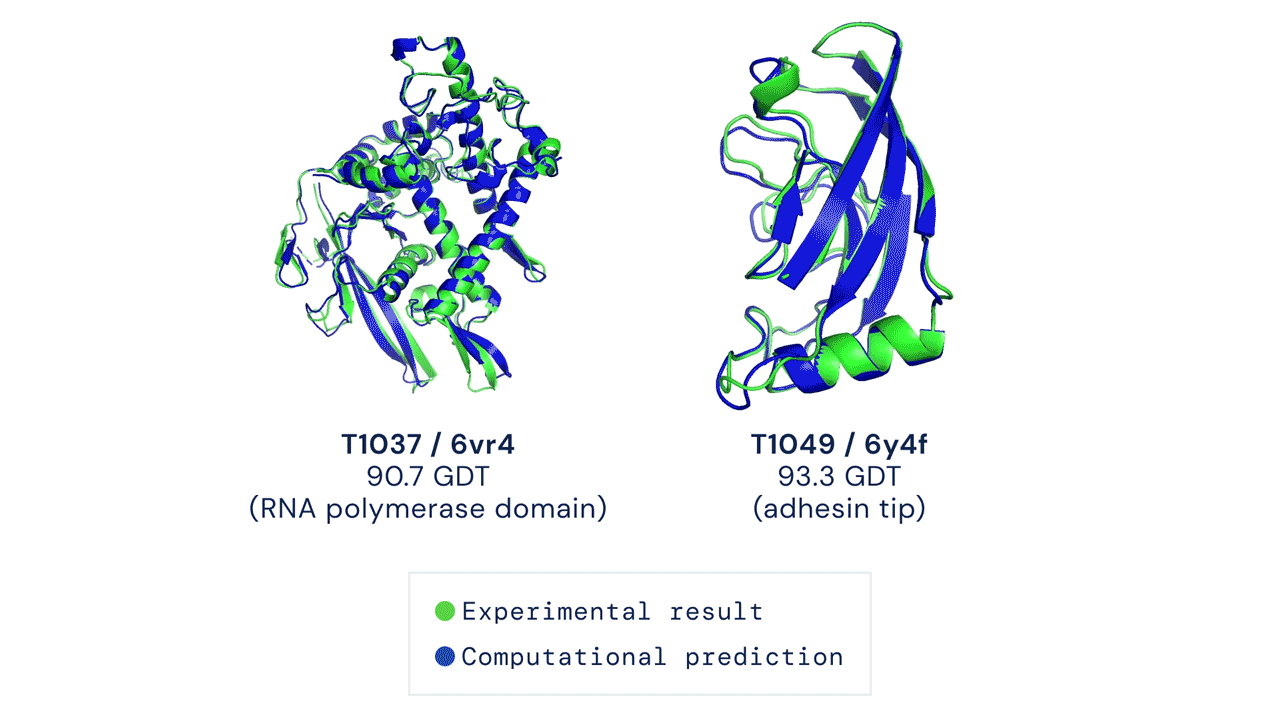
What’s with all the fuss?
This was only a challenge on which AlphaFold won with their technology…but can we expect this to be used in science and in improving our lives? According to all the reports, yes!
The first application in medicine is developing drugs faster and more precise. Drug development takes more than 10 years and billions of dollars, but systems such as AlphaFold have the potential to make this more efficient. Knowing the exact structure of a target protein helps, right?
Another application in medicine is understanding disease processes. The system allows us to identify almost exactly what a protein looks like and how it interacts with our cells. This might be a big advantage in understanding diseases such as Alzheimer’s or Huntington’s disease and ALS for example.
Finally, they also stress that it could help us find proteins and enzymes, perhaps even develop them, that break down plastic. Or maybe even capture carbon from the atmosphere. Along with medicine, it could revolutionise our fight against climate change and give us a new tool into our toolbox. It’s an awesome application, but that cannot make us more irresponsible.
If we look back, there have been thousands of ground-breaking discoveries that we all thought are going to revolutionise medicine and science. This is great news, but we also need to actively implement this and use it to our advantage. Until then, it's just an extra thing we can do, but don’t actually do it.





